July 2023
An atlas of healthy and injured cell states and niches in the human kidney.
July 2023
Single-cell Profiling of Prurigo Nodularis Demonstrates Immune-Stromal Crosstalk Driving Profibrotic Responses and Reversal with Nemolizumab.
July 2023
SiftCell: A robust framework to detect and isolate cell-containing droplets from single-cell RNA sequence reads.
July 2023
Pansclerotic morphea is characterized by IFN-γ responses priming dendritic cellfibroblast crosstalk to promote fibrosis.
July 2023
Genetic control of the dynamic transcriptional response to immune stimuli and glucocorticoids at single-cell resolution.
June 2023
Imaging-Based Screening Identifies Modulators of the eIF3 Translation Initiation Factor Complex in Candida albicans.
June 2023
Differentiation of IL-26+ TH17 intermediates into IL-17A producers via epithelial crosstalk in psoriasis.
June 2023
Analysis of Donor Pancreata Defines the Transcriptomic Signature and Microenvironment of Early Neoplastic Lesions.
March 2023
Multi-omic single-cell velocity models epigenome-transcriptome interactions and improves cell fate prediction.
February 2023
Subclonal evolution and expansion of spatially distinct THY1-positive cells is associated with recurrence in glioblastoma.
February 2023
Bone marrow mesenchymal stem cells induce metabolic plasticity in estrogen receptor-positive breast cancer.
January 2023
Two-sample test with g-modeling and its applications.
January 2023
SGLT2 inhibitors mitigate kidney tubular metabolic and mTORC1 perturbations in youth onset type 2 diabetes.
January 2023
Subclonal evolution and expansion of spatially distinct THY1-positive cells is associated with recurrence in glioblastoma.
January 2023
Recording of cellular physiological histories along optically readable self-assembling protein chains.
January 2023
Single-cell transcriptomics reveals a mechanosensitive injury signaling pathway in early diabetic nephropathy.
January 2023
WNT signaling in the tumor microenvironment promotes immunosuppression in murine pancreatic cancer.
December 2022
Computational modeling implicates protein scaffolding in p38 regulation of Akt.
December 2022
HIF1α lactylation enhances KIAA1199 transcription to promote angiogenesis and vasculogenic mimicry in prostate cancer.
December 2022
Single-cell RNA-sequencing identifies anti-cancer immune phenotypes in the early lung metastatic niche during breast cancer.
November 2022
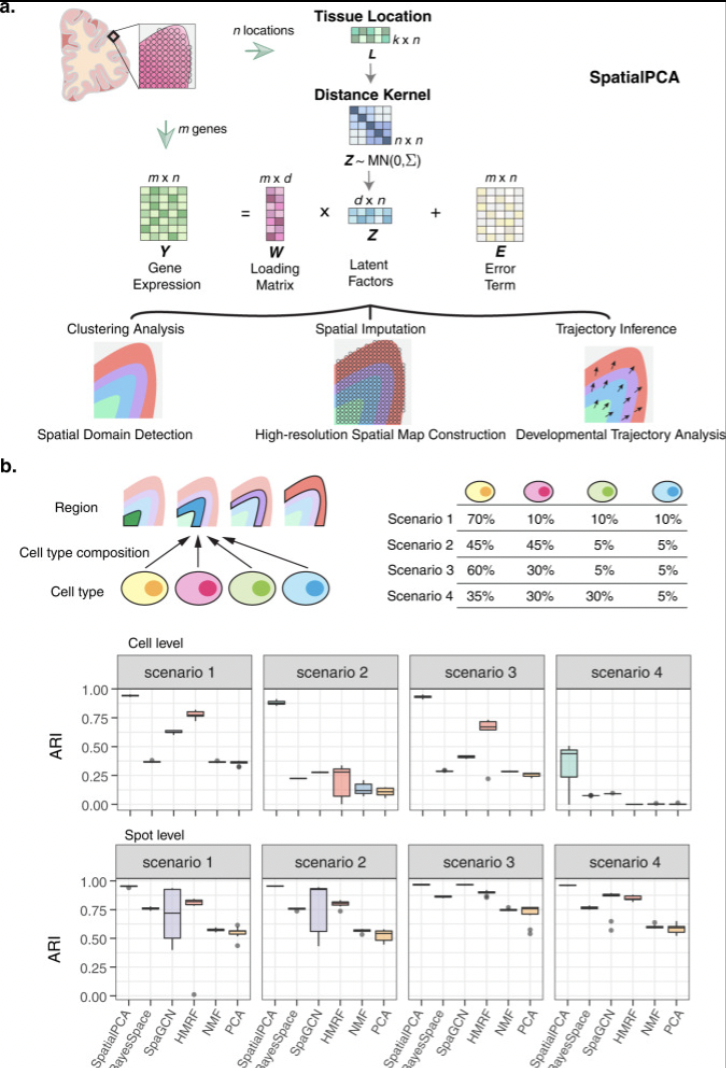
Spatially aware dimension reduction for spatial transcriptomics.
November 2022
The fate of early perichondrial cells in developing bones.
September 2022
Keap1 moderates the transcription of virus induced genes through G9a-GLPand NFκB p50 recruitment.
August 2022
BASS: multi-scale and multi-sample analysis enables accurate cell type clustering and spatial domain detection in spatial transcriptomic studies.
August 2022
Dissecting the Heterogeneity of Human Thoracic Aortic Aneurysms Using Single-Cell Transcriptomics.
July 2022
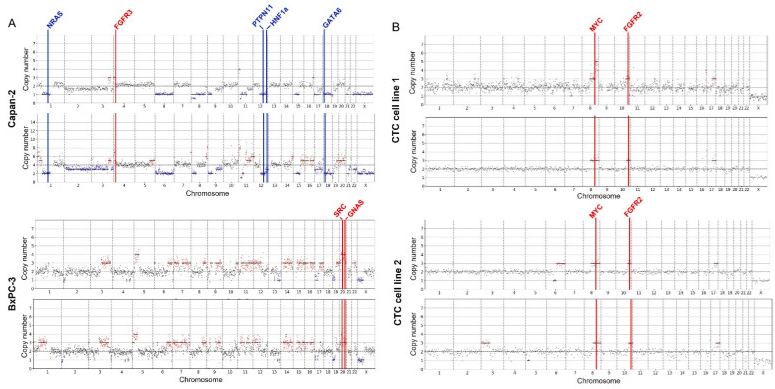
Integrated Workflow for the Label-Free Isolation and Genomic Analysis of Single Circulating Tumor Cells in Pancreatic Cancer.
July 2022
Spatial Transcriptomics as a Novel Approach to Redefine Electrical Stimulation Safety.
July 2022
Statistical analysis of spatially resolved transcriptomic data by incorporating multiomics auxiliary information
July 2022
Joint dimension reduction and clustering analysis of single-cell RNA-seq and spatial transcriptomics data.
September 2021